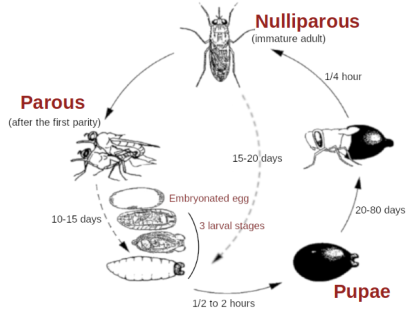
This program is designed to simulate the spatio-temporal dynamics of tsetse fly populations in a deterministic manner. The species studied here is Glossina palpalis gambiensis, in the Niayes region of Senegal.
The space is subdivided into cells of 250m x 250m resolution. Environmental heterogeneity is taken into account by the including temperature, which varies in both time and space, and carrying capacity, variable in space and constant in time. Based on daily temperature grids and a single carrying capacity grid, the program predicts the evolution of fly numbers per cell and their dispersion in the landscape. It is a compartmental model. The stages of development represented are pupae (males and females combined), young males, breeding males, nulliparous females, adult females of parity 1 to 4+. The model is deterministic, mechanistic, and the different parameters of the mortality, development and dispersion equations can be modified by the user, either on the command line or read from a file. Several aggregated outputs can be generated at the end of the simulation and stored in a text file, or put in the form of graphs for some of them. Initial conditions can be read from files (from the end of a previous simulation for example).
The program can also simulate control scenarios, based on increased mortality in certain cells, and to optimize the choice of cells to be treated and the level of treatment to be applied to achieve a given objective.
Associated publication: Cecilia H, Arnoux S, Picault S, Dicko A, Seck MT, Sall B, Bassène M, Vreysen M, Pagabeleguem S, Bancé A, Bouyer J and Ezanno P. Environmental heterogeneity drives tsetse fly population dynamics and control. bioRxiv 493650., ver. 3 peer-reviewed and recommended by PCI Ecology (2019).
The program is available on https://sourcesup.renater.fr/projects/spatial-tsetse/ or directly with the following command: git clone https://git.renater.fr/anonscm/git/spatial-tsetse/spatial-tsetse.git
For further information, please contact Sébastien Picault
This tool has been developed within the Dynamo team, in charge of modeling the pathogen propagation in animal populations.



