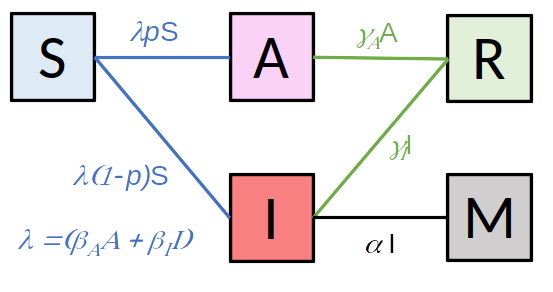
BIOEPAR is developing simulation models to predict the spread of pathogens under a wide range of contrasting situations and at different scales (batch of animals, herd, region, production chain), as well as the population dynamics of their vectors where appropriate. These models make it possible to better target surveillance strategies for the (re-)emergence of animal diseases, and to prioritise interventions to limit the socio-economic impact, and the impact on animal health and welfare, of the spread of deseases25. Some models take into account the decision-making process of health managers (farmers, groups, public decision-makers).
More specifically, we are developing software suites and mechanistic models, often stochastic, applied to various animal diseases or vectors:
* Cross-disciplinary software and algorithms
- EMULSION49.
- PASTE47.
- Network propagation & movement guidance based on epidemiological risk44
- Transmissio: data visualization:
- bayesian inference R package BRREWABC « Batched Resilient and Rapid Estimation Workflow through Approximate Bayesian Computation »
* Models on a regional scale / production chain
- between cattle herds:
BVD51
FCO14
FQ46
PTB4,5,23
Algorithms for organising the movement of young cattle between breeder and fattener farms43, 45
- between pig herds: Salmonnelle portage27,38
- multi-species hosts:
FCO15
FVR9,12
PPA2,26,50: ASF Challenge 2020 ; ASF Challenge INRAE team
- in aquatic environments: Vibrio aestuarianus in a spatialised oyster population36
* Herd-scale models
- cattle herd:
BVD1,21,22: Within herd model ; Farmer decisions
FQ16-18
PTB6,8,40_42
Genetic selection
DHM29,30
- pig herd :
SDRP
Salmonnelle portage34
- wildlife: pestivirus in a population of isards3,35
* Lot-scale models
- batch of young cattle: BRD48.52
- Vibrio aestuarianus in an oyster population in an aquarium37
* Models on an intra-host scale
- macrophage immune response to PRRS virus infection28
- intra-host and intra-vector infection dynamics of RVF virus13
- intra-mosquito viral infection dynamics (in progress)
- lymph node response to antigen induction (ongoing)
* Decision models
- coupling epidemiological models and individual intervention decisions: generic framework19; application to BVD21
- collective decision: management of PRRS54; generic framework20
* Vector population dynamics models
- Mosquitoes7,24,53
- Tsetse10,11
- Ticks34 and diseases transmitted: bovine babesiosis33 and CCHF31-33
[Disease abbreviations : BRD = bovine respiratory disease, BVD = bovine viral diarrhoea, CCHF = Crimean-Congo haemorrhagic fever, Bluetongue = bluetongue, QF = Q fever, RVF = Rift Valley fever, ASF = African swine fever, BTP = bovine paratuberculosis, PRRS = porcine respiratory dysgenic syndrome]
If you are interested in any of these models, please contact Sébastien Picault
References
- Arnoux S., Bidan F., Damman A., Petit E., Assié S., Ezanno P. 2021. To vaccinate or not: impact of bovine viral diarrhoea in French cow-calf herds. Vaccines 9(10), 1137. https://doi.org/10.3390/vaccines9101137
- Beaunée G., Deslandes F., Vergu E., 2023. Inferring ASF transmission in domestic pigs and wild boars using a paired model iterative approach. Epidemics 42, 100665. https://doi.org/10.1016/j.epidem.2023.100665
- Beaunée G., Gilot-Fromont E., Garel M., Ezanno P. 2015. Seasonal spread of a Pestivirus in a structured Pyrenean chamois population. Vet Res 46(1):86, doi:10.1186/s13567-015-0218-8
- Beaunée G., Vergu E., Ezanno P. 2015. Modelling of paratuberculosis spread between dairy cattle farms at a regional scale. Vet Res 46:111, doi:10.1186/s13567-015-0247-3
- Beaunée G., Vergu E., Joly A., Ezanno P. 2017. Controlling bovine paratuberculosis at a regional scale: towards a decision modeling tool. J Theor Biol 435:157-183, doi:10.1016/j.jtbi.2017.09.012
- Ben Romdhane R., Beaunée G., Camanes G., Guatteo R., Fourichon C., Ezanno P. 2017. Which phenotypic traits of resistance should be improved in cattle to control paratuberculosis dynamics in a dairy herd: a modelling approach. Vet Res 48:62, doi:10.1186/s13567-017-0468-8
- Cailly P., Tran A., Balenghien T., L’Ambert G., Toty C., Ezanno P. 2012. A climate-driven abundance model to assess mosquito control strategies. Ecol Model 227, 7-17, doi:10.1016/j.ecolmodel.2011.10.027
- Camanes G., Joly A., Fourichon C., Ben Romdhane R., Ezanno P. 2018. Control measures to avoid increase of paratuberculosis prevalence in dairy cattle herds: an individual-based modelling approach. Vet Res 49:60, https://doi.org/10.1186/s13567-018-0557-3
- Cavalerie L.*, Charron M.V.P.*, Ezanno P.*, Dommergues L., Zumbo B., Cardinale E. 2015. A stochastic model to study Rift Valley Fever persistence with different seasonal patterns of vector abundance: new insights on the endemicity in the tropical island of Mayotte. PLoS ONE 10(7):e0130838, doi:10.1371/journal.pone.0130838
- Cecilia H., Arnoux S., Picault S., Dicko A., Seck M.T., Sall B., Bassène M., Vreysen M., Pagabeleguem S., Bancé A., Bouyer J., Ezanno P. 2019. Environmental heterogeneity drives tsetse fly population dynamics and control. bioRxiv, https://doi.org/10.1101/493650, ver. 3 peer-reviewed and recommended by PCI Ecology (https://dx.doi.org/10.24072/pci.ecology.100024).
- Cecilia H., Arnoux S., Picault S., Dicko A., Seck M.T., Sall B., Bassène M., Vreysen M., Pagabeleguem S., Bancé A., Bouyer J., Ezanno P. 2021. Dispersal in heterogeneous environments drives population dynamics and control of tsetse flies. Proceedings of the Royal Society B 20202810. https://doi.org/10.1098/rspb.2020.2810
- Cecilia H., Métras R., Fall A.G., Lo M., Lancelot R., Ezanno P. 2020. It’s risky to wander in September: modelling the epidemic potential of Rift valley fever in a Sahelian setting. Epidemics 33, 100409, https://doi.org/10.1016/j.epidem.2020.100409
- Cecilia H., Vriens R., Schreur P.W., de Wit M., Métras R., Ezanno P., ten Bosch Q. Heterogeneity of Rift valley fever virus transmission potential across livestock hosts, quantified through a model-based analysis of host viral load and vector infection. PLoS Computational Biology 18(7): e1010314. https://doi.org/10.1371/journal.pcbi.1010314
- Charron M., Langlais M., Seegers H., Ezanno P. 2011. Seasonal spread and control of Bluetongue in cattle. J Theor Biol 291, 1-9, doi:10.1016/j.jtbi.2011.08.041
- Charron M.V.P., Kluiters G., Langlais M., Seegers H., Baylis M., Ezanno P. 2013. Seasonal and spatial heterogeneities in host and vector abundances impact the spatiotemporal spread of bluetongue. Vet Res 44:44, doi:10.1186/1297-9716-44-44
- Courcoul A., Hogerwerf L., Klinkenberg D., Nielen M., Vergu E., Beaudeau F., 2011. Modelling effectiveness of herd level vaccination against Q fever in dairy cattle. Vet. Res., 42(1):68.
- Courcoul A., Monod H., Nielen M., Klinkenberg D., Hogerwerf L., Beaudeau F., Vergu E., 2011. Modelling the effect of heterogeneity of shedding on the within herd Coxiella burnetii spread and identification of key parameters by sensitivity analysis. J. Theor. Biol., 284(1):130-141.
- Courcoul A., Vergu E., Denis J.-B., Beaudeau F., 2010. Spread of Q fever within dairy cattle herds: key parameters inferred using a Bayesian approach. Proceedings of the Royal Society B: Biological Sciences, 277(1695):2857-2865.
- Cristancho Fajardo L., Ezanno P., Vergu E. 2021. Integrative modelling of pathogen spread through animal trade by accounting for farmers’ control decisions. Scientific Report 11:9581 https://doi.org/10.1038/s41598-021-88471-6
- Cristancho L., Ezanno P., Vergu E. 2022. Dynamic resource allocation for controlling pathogen spread on a large animal metapopulation network. Journal of the Royal Society Interface 19: 20210744. https://doi.org/10.1098/rsif.2021.0744
- Cristancho-Fajardo L., Vergu E., Beaunée G., Arnoux S., Ezanno P. 2022. Learning and strategic imitation in modelling farmers’ dynamic decisions on Bovine Viral Diarrhoea vaccination. Veterinary Research 53:102. https://doi.org/10.1186/s13567-022-01112-2
- Damman A., Viet A-F, Arnoux S., Guerrier-Chatellet M-C, Petit E, Ezanno P. 2015. Modeling the spread of bovine viral diarrhea virus (BVDV) in a beef cattle herd and its impact on herd productivity. Vet Res 46:12, doi:10.1186/s13567-015-0145-8
- Ezanno P., Arnoux S., Joly A., Vermesse R. 2022. Rewiring cattle trade movements helps to control bovine paratuberculosis at a regional scale. Preventive Veterinary Medicine 198:105529. https://doi.org/10.1016/j.prevetmed.2021.105529
- Ezanno P., Balenghien T., Arnoux S., Cailly P., Aubry-Kientz M., L’Ambert G., Toty C., Tran A. 2015. A generic climate-driven model to predict mosquito population dynamics and the associated risk of mosquito-borne disease transmission. Prev Vet Med 120(1), 39-50, doi:10.1016/j.prevetmed.2014.12.018
- Ezanno P., Beaunée G., Picault S., Arnoux S., Sicard V., Beaudeau F., Rault A., Vergu E. 2018. Gestion des maladies endémiques du troupeau aux territoires : contribution de la modélisation épidémiologique pour soutenir la prise de décision (projet MIHMES, 2012-2017). Innovations Agronomiques 68, 53-65.
- Ezanno P., Picault S., Bareille S., Beaunée G., Boender G.J., Dankwa E.A., Deslandes F., Donnelly C.A., Hagenaars T.J., Hayes S., Jori F., Lambert S., Mancini M., Munoz F., Pleydell D.R.J., Thompson R.N., Vergu E., Vignes M., Vergne T. 2022. The African swine fever modelling challenge: model comparison and lessons learnt. Epidemics 40, 100615. https://doi.org/10.1016/j.epidem.2022.100615
- Ferrer Savall J., Bidot, C., Leblanc-Maridor M., Belloc C., Touzeau S. 2016 Modelling Salmonella transmission among pigs from farm to slaughterhouse: interplay between management variability and epidemiological uncertainty. Internal J Food Microbiol 229:33–43, doi:10.1016/j.ijfoodmicro.2016.03.020
- Go N., Bidot C., Belloc C., Touzeau S., 2014. Integrative model of the immune response to a pulmonary macrophage infection: what determines the infection duration? PLoS ONE, 9(9):e107818, doi:10.1371/journal.pone.0107818
- Gontier P., Bareille N. 2023. Description fonctionnelle et modalités d'emploi. INRAE. doi:10.17180/NPMC-KH96
- Gontier P., Bareille N., Picault S., 2022. Dairy Health Manager : un simulateur multi-agents flexible pour l'étude des maladies des animaux d'élevage. Journées Francophones sur les Systèmes Multi-Agents (JFSMA), Plate-Forme Intelligence Artificielle (PFIA). hal-03710369.
- Hoch T., Breton E., Josse M., Deniz A., Guven E., Vatansever Z. 2016. Identifying main drivers and testing control strategies for CCHFV spread. Exp Appl Acarol 68(3):347-359.
- Hoch T., Breton E., Vatansever Z. 2018. Dynamic Modeling of Crimean Congo Hemorrhagic Fever Virus (CCHFV) Spread to Test Control Strategies. J Med Entomol. 55:1124-1132. DOI: 10.1093/jme/tjy035
- Hoch T., Goebel J., Agoulon A., Malandrin L. 2012. Modelling bovine babesiosis: A tool to simulate scenarios for pathogen spread and to test control measures for the disease. Prev Vet Med 106:136-142.
- Hoch T., Monnet Y., Agoulon A. 2010. Influence of host migration between woodland and pasture on the population dynamics of the tick Ixodes ricinus: A modelling approach. Ecol Model 221:1798-1806.
- Lambert S., Ezanno P., Garel M., Gilot-Fromont E. 2018. Stochasticity drives epidemiological patterns in wildlife with implications for diseases and population management. Sci Rep 8:16846. doi:10.1038/s41598-018-34623-0
- Lupo C., Dutta B.L., Petton S., Ezanno P., Tourbiez D., Travers M-A., Pernet F., Bacher C. 2020. Spatial epidemiological modelling of infection by Vibrio aestuarianus shows that connectivity and temperature control oyster mortality. Aquaculture Environment Interactions 12, 511-527, https://doi.org/10.3354/aei00379
- Lupo C., Travers M-A., Tourbiez D., Barthélémy C.F., Beaunée G., Ezanno P. 2019. Modeling the transmission of Vibrio aestuarianus in Pacific oysters using experimental infection data. Front Vet Sci 6:142, https://doi.org/10.3389/fvets.2019.00142
- Lurette A., Belloc C., Keeling M., 2011. Contact structure and Salmonella control in the network of pig movements in France. Prev. Vet. Med., 102(1):30-40.
- Lurette A., Touzeau S., Ezanno P., Hoch T., Seegers H., Fourichon C., Belloc C. 2011. Within-herd biosecurity and Salmonella seroprevalence in slaughter pigs: a simulation study. J Anim Sci 89, 2210-2219, doi:jas.2010-2916v1-20102916
- Marcé C., Ezanno P., Seegers H., Pfeiffer D., Fourichon C. 2011. Predicting fadeout versus persistence of paratuberculosis in a dairy cattle herd for management and control purposes: a modelling study. Vet Res 42:36, doi:10.1186/1297-9716-42-36
- Marcé C., Ezanno P., Seegers H., Pfeiffer D., Fourichon C. 2011. Within-herd contact structure and transmission of Mycobacterium avium subspecies paratuberculosis in a persistently infected dairy cattle herd. Prev Vet Med 100, 116-125, doi:10.1016/j.prevetmed.2011.02.004
- More S., Cameron A., Strain S., Cashman B., Ezanno P., Kenny K., Fourichon C., Graham D. 2015. Evaluation of testing strategies to identify infected animals at a single round of testing within dairy herds known to be infected with Mycobacterium avium subsp. paratuberculosis. J. Dairy Sci 98:5194–5210
- Morel-Journel T., Assié S., Vergu E., Mercier J.-B., Bonnet-Beaugrand F., Ezanno P. 2021. Minimizing the number of origins in batches of weaned calves to reduce their risks of developing bovine respiratory diseases. Veterinary Research 52:5, https://doi.org/10.1186/s13567-020-00872-z
- Morel-Journel T., Ezanno P., Vergu E. Rewiring cattle movements to limit infection spread. Veterinary Research (accepted July 2024), preprint: https://doi.org/10.1101/2022.08.24.505123
- Morel-Journel T., Mercier J-B., Bareille N., Vergu E., Ezanno P. 2021. Selecting sorting centres to avoid long distance transport of weaned beef calves. Scientific Report 11, 1289. https://doi.org/10.1038/s41598-020-79844-4
- Pandit P., Hoch T.*, Ezanno P.*, Beaudeau F., Vergu E. 2016. Q fever spread between dairy cattle herds in an enzootic region: modelling contributions of airborne transmission and trade. Vet Res 47:48, doi:10.1186/s13567-016-0330-4
- Picault P., Niang G., Sicard V., Sorin-Dupont B., Assié S., Ezanno P. 2024. Leveraging artificial intelligence and software engineering methods in epidemiology for the co-creation of decision-support tools based on mechanistic models. Preventive Veterinary Medicine - special issue: Artificial Intelligence in Veterinary Epidemiology 228:106233. https://doi.org/10.1016/j.prevetmed.2024.106233; https://hal.inrae.fr/hal-04593328 (preprint: https://biorxiv.org/cgi/content/short/2023.09.03.555060v1)
- Picault S., Ezanno P., Smith K., Amrine D., White B., Assié S. 2022. Modelling the effects of antimicrobial metaphylaxis and pen size on bovine respiratory disease in high and low risk fattening cattle. Veterinary Research 53:77. https://doi.org/10.1186/s13567-022-01094-1
- Picault S., Huang Y.-L., Sicard V., Arnoux S., Beaunée G., Ezanno P. 2019. EMULSION: transparent and flexible multiscale stochastic models in human, animal and plant epidemiology. PLoS Comput Biol 15(9): e1007342, https://doi.org/10.1371/journal.pcbi.1007342
- Picault S., Vergne T., Mancini M., Bareille S., Ezanno P. 2022. The African swine fever modelling challenge: objectives, model description and synthetic data. Epidemics 40, 100616. https://doi.org/10.1016/j.epidem.2022.100616
- Qi L., Beaunée G., Arnoux S., Dutta B.L., Joly A., Vergu E., Ezanno P. 2019. Neighbourhood contacts and trade movements drive the regional spread of bovine viral diarrhoea virus (BVDV). Vet Res 50:30, https://doi.org/10.1186/s13567-019-0647-x
- Sorin-Dupont B., Picault S., Pardon B., Ezanno P.*, Assié S.* 2023. Modeling the effects of farming practices on bovine respiratory disease in a multi-batch cattle fattening farm. Preventive Veterinary Medicine 106009. [*Contribution équivalente] https://doi.org/10.1016/j.prevetmed.2023.106009
- Tran A., L'Ambert G., Lacour G., Benoît R., Demarchi M., Cros M., Cailly P., Aubry-Kientz M., Balenghien T., Ezanno P. 2013. A rainfall- and temperature-driven abundance model for Aedes albopictus populations. Internal J Envir Res Public Health, 10(5), 1698-719, doi:10.3390/ijerph10051698
- Viet A-F., Krebs S., Rat-Aspert O., Jeanpierre L., Belloc C., Ezanno P. 2018. A modelling framework based on MDP to coordinate farmers' disease control decisions at a regional scale. PLoS ONE 13(6): e0197612, doi:10.1371/journal.pone.0197612



